T3: Large-scale data fusion by collective matrix factorization
Organizers:
Marinka Zitnik (University of Ljubljana, Slovenia) and Blaz Zupan (University of Ljubljana, Slovenia & Baylor College of Medicine, Houston, USA)
Tutorial Summary:
Motivation
We are overwhelmed with data of different types and sizes. In everyday life, we prefer to make decisions by considering all the available information, and often find that the inclusion of even seemingly circumstantial evidence provides an advantage. It would be great if our computational methods could conjecture in a similar way. In molecular biology, incorporating all the data, especially indirect information, may provide substantial gains in the accuracy of predictions and help to deepen our overall understanding of the problem data.
In this tutorial, we will present a high-level introduction to biomedical data integration and data fusion, highlighting the state of the art in the field as well as outstanding computational challenges. These include an introduction to the goals of typical data fusion studies and the bioinformatics techniques currently available to achieve them. We will then focus on matrix factorization-based data fusion approaches. We will introduce a technique for fusing any number of data sets that is agnostic to any specific model organism or data domain, discuss our own work on large-scale data fusion, and demonstrate our data fusion toolbox on several studies. The tutorial will include an easy-to-follow explanation of relevant mathematical concepts and a showcase with benchmarks and examples that attendees will be able to reproduce during the tutorial through either programmatic access to our toolbox or via tinkering in a user-friendly visual programming environment.
Data fusion approaches that will be discussed in detail in the tutorial build upon latent models and matrix factorization to collectively address possibly large volumes of heterogeneous data and construct accurate prediction models. In particular, for problems with heterogeneity in both the prediction task and data, we have recently proposed a data mining approach that can jointly learn in multiple related tasks with overlapping, partially overlapping or even completely different feature spaces. The factorization-based data fusion leaves the data in their own domain spaces, requires no or minimal data engineering at the input, and is able to leverage heterogeneous data to improve learning performance. Just like with Lego bricks, the approach assembles the data in a mosaic called a data fusion graph that through joint compression of all data matrices yields a model wherein every piece of evidence counts, even if distantly related to the prediction task. The method we will present can be applied both to unsupervised and supervised modeling of the data, and provides a venue to jointly collectively consider a plethora of information available in molecular biology.
Expected Goals
The goal of the tutorial is to familiarize the audience with the concepts and methods of data fusion. The presentation of computational approaches will be accompanied with practical training using visual programming and scripting tools. By the end of the tutorial, the attendees should be able to execute data fusion-based analysis on their own data and use data fusion in functional genomics and gene prioritization. Our goals are to:
- Train the audience to identify situations where data fusion could yield substantial benefit over traditional data mining approaches.
- Learn how to prepare heterogeneous data for data fusion.
- Explain the mathematics behind data fusion algorithms, including concepts such as matrix tri-factorization, matrix completion, collective matrix factorization and latent factor chaining.
- Understand the inner workings of data fusion by experimenting with data fusion techniques within an easy-to-use visual programming and data visualization environment.
- Illustrate the programmatic access to our data fusion toolbox.
- Demonstrate the utility of data fusion in functional genomics, gene prioritization, association mining and clustering.
- Provide guidelines that will help attendees to recognize situations where data fusion may be appropriate and would be expected to yield good predictive performance.
- Through brainstorming, identify the many potential applications of data fusion.
To achieve these goals we will intertwine short lectures with hands-on activities, where attendees will have the opportunity to work with our data fusion software and explore the benefits of data fusion in a series of real-world examples.
Level Intermediate.
A basic knowledge of data mining and omics is assumed. We will make every effort to explain data fusion intuitively and in simple terms, thus trying to reach out to both biologists and computational scientists. We will present the mathematical background of the data fusion methodology, guide the hands-on parts of the tutorial, and use Python programming in a short section of tutorial, taking care not to be too technical and with the aim of motivating the computational scientists in the audience to explore further.
Intended Audience Computational scientists, data mining researchers, and molecular biologists with interest in data analysis and large-scale data integration.
Prerequisites Attendees are encouraged to bring their own laptops to the tutorial and to install our visual programming software prior to the event. A special installation of Orange data mining suite will be prepared for the tutorial and will be accompanied with the data needed to perform the case studies.
Tutorial Agenda:
Tuesday, June 9, 2015
| 14:00 – 15:30 |
Introduction
|
| 15:30 – 16:00 | Coffee break. |
| 16:00 – 17:30 |
Data fusion with case studies
|
| 17:30 – 17:45 |
Discussion and conclusions
|
Tutorial speakers:
Blaz Zupan is professor of computer science at the University of Ljubljana, and a visiting professor at Baylor College of Medicine in Houston. His research focuses on methods for data mining and applications in bioinformatics and systems biology. Marinka Zitnik is currently working towards her PhD in computer science. Her research interests include machine learning, optimization and matrix analysis.
Blaz Zupan is coauthor of Orange (http://orange.biolab.si), a Python-based and visual programming data mining suite, and several bioinformatics web applications, such as dictyExpress for gene expression analytics (http://dictyexpress.biolab.si) and GenePath for epistasis analysis (http://genepath.biolab.si).
Together with Marinka Zitnik, they developed Nimfa, a Python programming library for nonnegative matrix factorization (http://nimfa.biolab.si). They have recently coorganized several workshops (http://goo.gl/kt1Ao9, http://goo.gl/7EdjFp) with a few hundred audience members about techniques for large-scale functional genomics data analysis in Orange. Workshops included lectures and hands-on activities with practical exercises in data mining for researchers from the University of Ljubljana, Baylor College of Medicine, Houston, and the University of Toronto.
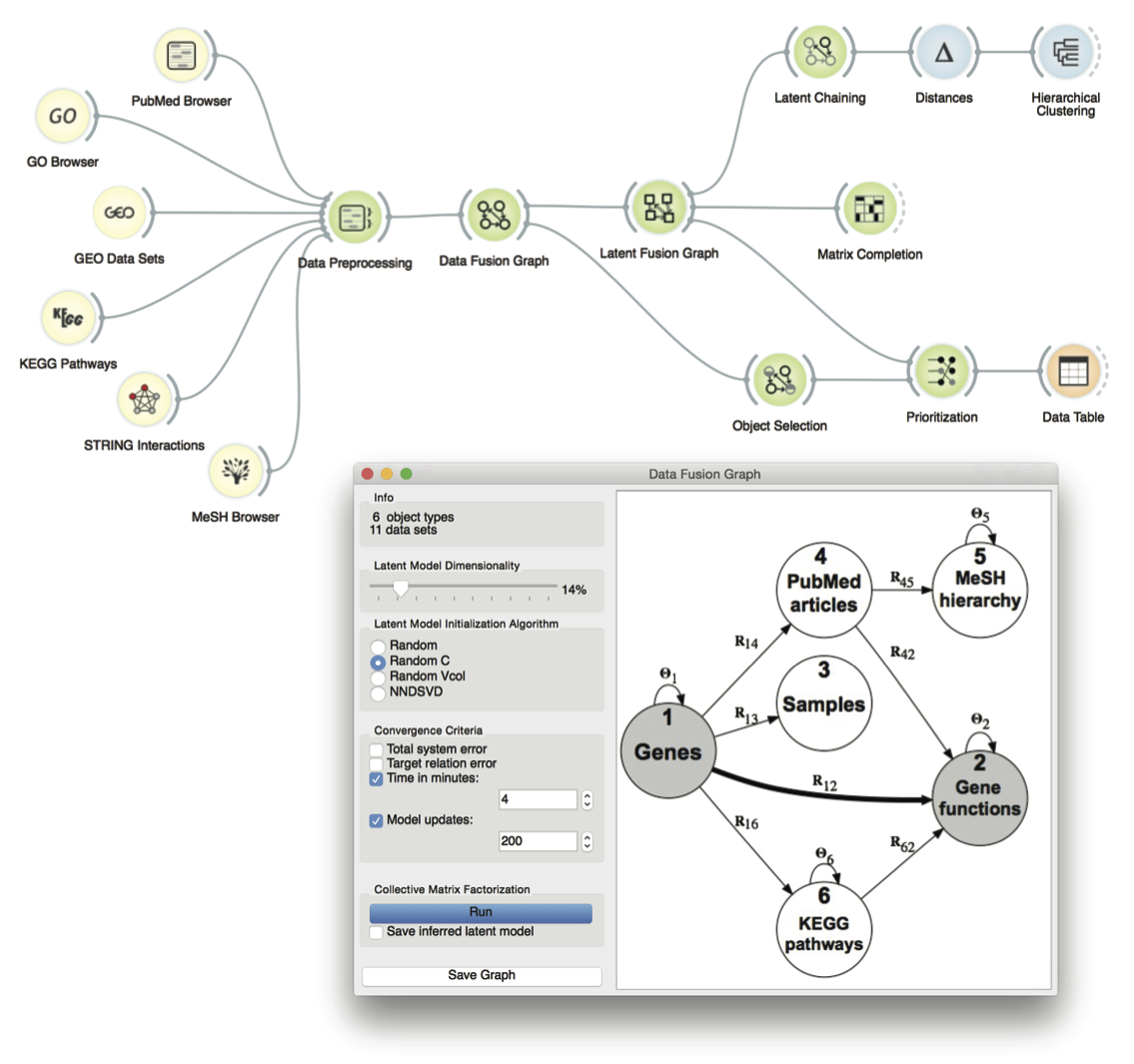
Figure: User interface of a data fusion toolbox that is implemented within the Orange data mining suite. Tutorial attendees will design, adapt and visualize workflows that will perform different prediction tasks, such as gene prioritization, clustering, association mining and matrix completion, all by fusing tens of different sources of biological information.






