T1: Computer-aided drug design with SwissDrugDesign
Organizers:
Vincent Zoete, Antoine Daina, Ute Röhrig, Olivier Michielin (SIB, University of Lausanne)
Tutorial Summary:
The attendee will be introduced to the latest developments in computer-aided drug design proposed by the Molecular Modeling Group of the SIB Swiss Institute of Bioinformatics. Using their own computer and a simple browser, attendees will predict secondary targets of small molecules in relation with toxicity issues or drug repurposing, estimate ADME properties, perform virtual high-throughput screening, and get proficient ideas about possible compound modifications based on the data mining of the medicinal chemistry literature.
Target audience: Everyone interested in drug design, discovery and development.
Prerequisites: Just bring your computer. The connection to the internet with a recent browser (chrome or Firefox preferably) is needed. Moreover the potential firewall must allow web services.
Tutorial Agenda:
Tuesday, September 12, 2017
Venue: Kollegienhaus, University of Basel
| 9:00 – 10:30 |
Session I - General introduction
|
| 10:30 – 11:00 | Coffee break |
| 11:00 – 12:30 |
Session II - Ligand-based virtual screening (theory and practice)
Session III - Target Prediction (theory and practice)
|
| 12:30 – 13:30 | Lunch break |
| 13:30 – 15:00 |
Session IV - ADME, pharmacokinetics, druglikeness (theory and practice)
|
| 15:00 – 15:30 | Coffee break |
| 15:30 – 17:00 |
Session V - Fragment-based drug design
|
Tutorial Speakers:
Vincent Zoete obtained an engineer’s degree in chemistry from the ENSCL (1995, Lille, France) with a DEA in Organic and Macromolecular chemistry, and holds a PhD in organic chemistry (1999). He joined SIB in 2004 as Research Manager for the Molecular Modeling Group (MMG), and became Associate Group Leader (2011). The group is developing freely accessible CADD web tools. The accent is put on the ease of use, to widen the access to nonexperts. Vincent is working on the development of protein engineering and drug design techniques, and collaborates extensively with different experimental research groups and pharma companies.
Several online tools were developed by MMG, such as:
- SwissDock (since 2010), a simple, yet professional docking web service to predict how of molecules bind to proteins.
- SwissParam (since 2010) provides topologies and parameters for the molecular modeling of small organic molecules.
- SwissBioisostere (since 2012) is the first comprehensive and freely accessible database collecting over 4.5 million molecular substructural replacements extracted from the literature, along with information on how frequently such replacements were performed in the past, and the observed impact on biological activity.
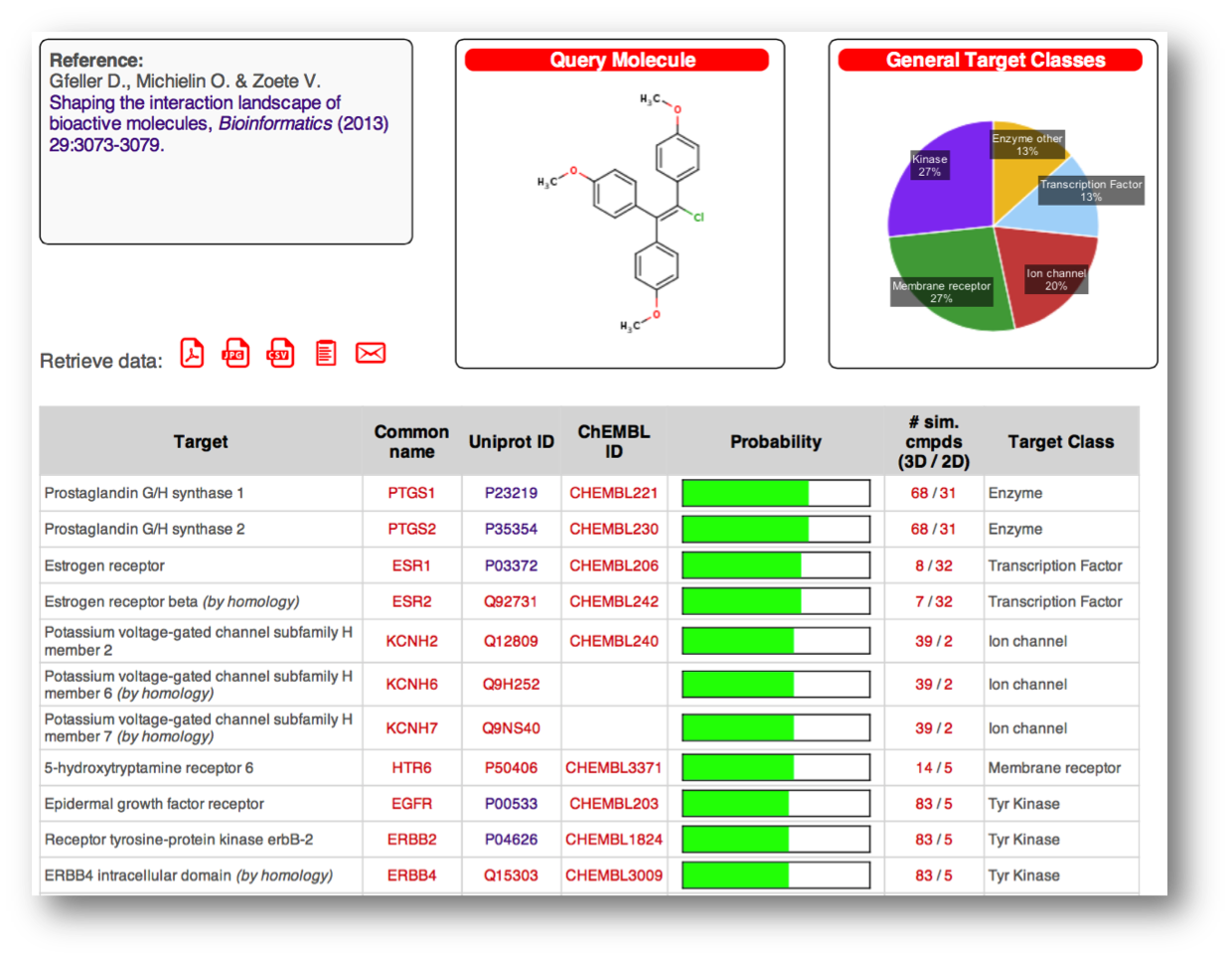
- SwissTargetPrediction (since 2014), a web service to predict the protein targets of bioactive small molecules, for toxicity prediction or drug repurposing.
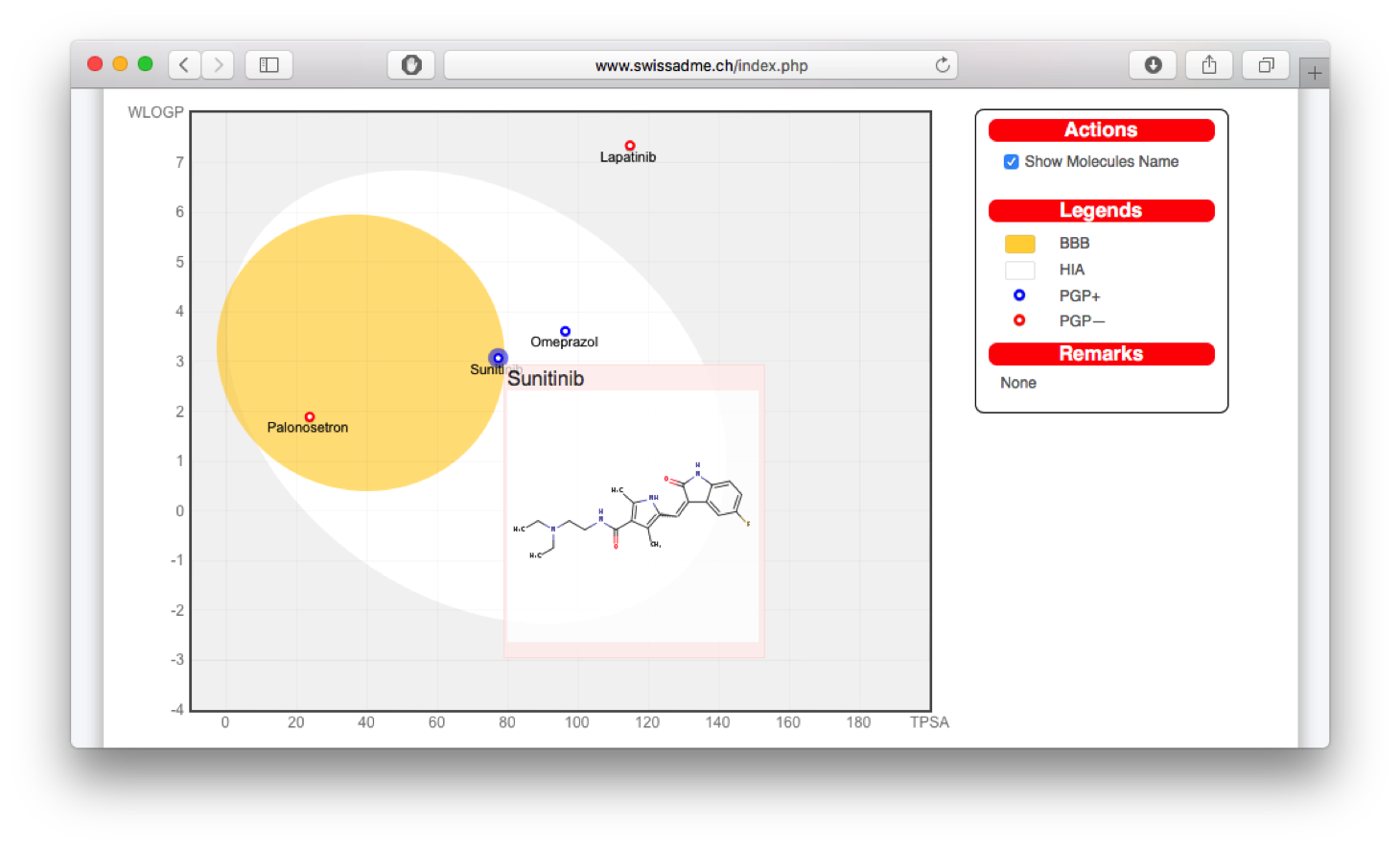
- SwissADME (since 2016), a web service to estimate physicochemical, pharmocokinetics and druglikeness properties.
- SwissSimilarity (since 2016) is the first online simple yet powerful, massive and fast ligand-?based virtual screening (VS). It includes collections of drugs, bioactive molecules, commercially available compounds and an unprecedented large virtual library of readily synthesizable compounds.
In the last 12 months, these web services received 105,000 unique visitors who submitted 110,000 jobs.
Vincent was co-Pi in the Agora DocKing project, made a major contribution to the development of the http://www.drug-design-workshop.ch web site, and was involved in outreach activities. He also teaches drug design and protein engineering for the SIB and the Universities of Lausanne, Geneva and Fribourg.
Antoine Daina is a pharmacist and got a PhD in Pharmaceutical Sciences (UNIGE, 2006). He spent 3 years as computational chemist at Syngenta, Chemistry Research (co-inventor of 4 patented bioactive molecular scaffolds). After 3 years at the School of Pharmaceutical Sciences, Geneva as Lecturer and Senior Scientist, he joined the MMG at SIB in 2012. Antoine’s academic focus is CADD, with an emphasis on pharmacokinetic models. His main teaching duties are drug design for Masters in Biochemistry and Doctoral students in Pharmacy. Antoine is a consultant for pharma companies (1 patent applied for clinical development compounds, 2016). He was involved in Agora DocKing to develop the web site and to conduct workshops.